
CHEN XunPh.D.
- Academic title:Principal Investigator
- Discipline:Computational Biology
- Departments:Multi-omics and AI research in infection and immunity
- Phone:86-21-54923180
- E-mail:xchen@siii.cas.cn
- Mailing Address:Life Science Research Building, 320 Yueyang Road, Xuhui District, Shanghai 200031, P.R. China
Academic experience
2024-present: Principal Investigator, Shanghai Institute of Immunity and Infection, CAS
2021-2024: Program-specific Assistant Professor, Kyoto University, Japan
2020-2021: Program-specific Researcher, Kyoto University, Japan
2014-2019: Postdoctoral researcher, University of Vermont, USA
Education
2008-2014: Ph.D., Huazhong Agricultural University, China
2005-2006: Minor, Huazhong University of Science and Technology, China
Human virome is complex and diverse, and associated with many human diseases. Influenza virus infection causes seasonal epidemics and results in a wide range of disease severity between individuals. Oncovirus infection is responsible for a large proportion of cancer in humans, while most of which could be integrated in the human genome. On the other hand, endogenous retrovirus, a subclass of transposable elements, are derived from ancient retroviruses. Through their distributed and enriched transcriptional factor bindings sites, they co-opt and re-shape the immune-related regulatory networks. Thus, endogenous retroviruses and other TEs play important roles in cancer, infection- and immune-related diseases.
Our lab focuses on developing innovative multi-omics sequencing technology, machine learning and bioinformatics algorithms to study the pathogenesis and individual variability of infection- and immune-related diseases, and to guide the development of vaccines and drugs for the prevention and precision medicine of these human diseases.
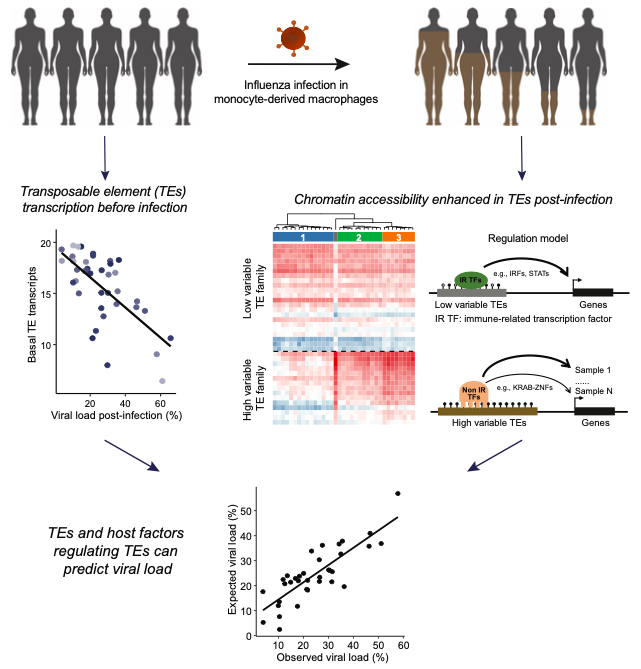
Figure 1. Transposable elements are associated with the inter-individual variation in immunity.
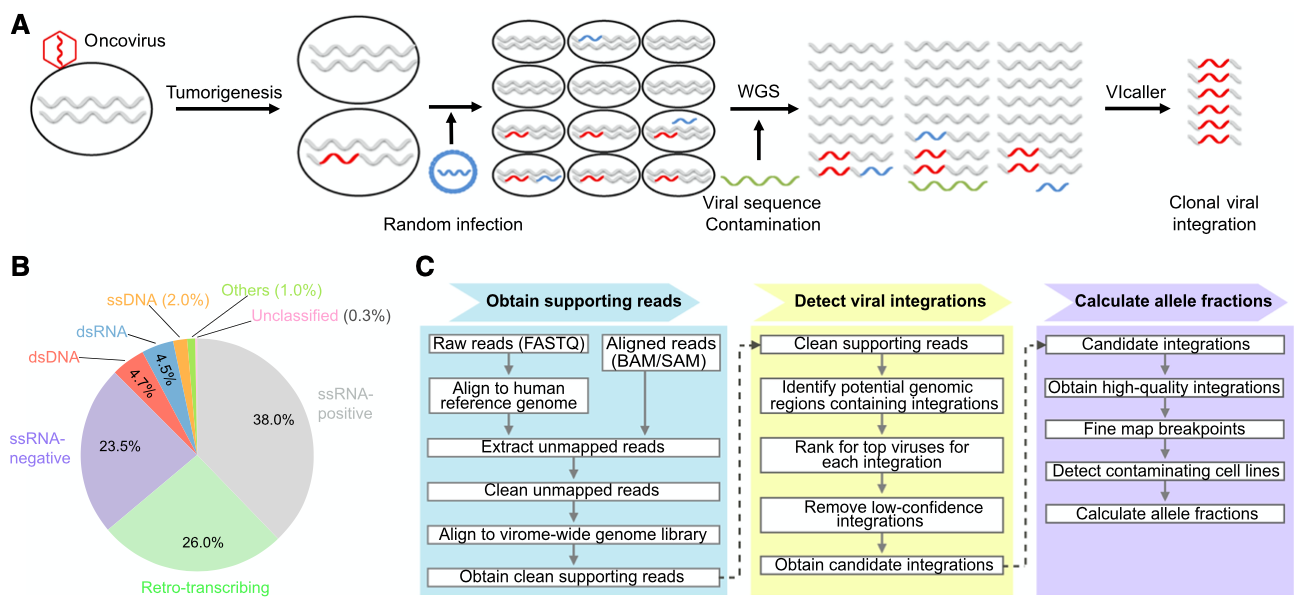
Figure 2. A virome-wide approach, VIcaller, to identify clonal viral elements (integrations) in early stages of tumorigenesis and novel oncoviruses.
Selected representative publications (#first-author,*corresponding-author):
1. Chen X#*, Zhang JC, Yan YZ, Bourque G*, Inoue F*: Cryptic endogenous retroviruses subfamilies in the primate lineage. BioRxiv. 2024, Under revision with Nature Communications (https://doi.org/10.1101/2023.12.07.570592)
2. Chen X#, Pacis A, Aracena A, Gona S, Kwan T, Groza C, Lin YL, Sindeaux R, Yotova V, Pramatarova A, Simon MM, Pastinen T, Barreiro L, Bourque G*: Transposable elements are associated with the variable response to influenza infection. Cell Genomics. 2023 May;3(5):100292 (https://doi.org/10.1016/j.xgen.2023.100292)
3. Chen X#, Kost J, Sulovari A, Wong N, Liang WS, Cao J, and Li D*: A virome-wide clonal integration analysis platform for discovering cancer viral etiologies. Genome Research. 2019 May;29(5):819-830 (https://genome.cshlp.org/content/29/5/819)
4. Chen X#, Li D*: ERVcaller: Identifying polymorphic endogenous retrovirus and other transposable element insertions using whole-genome sequencing data. Bioinformatics. 2019 Oct;35(20):3913-3922 (https://doi.org/10.1093/bioinformatics/btz205)

